FHIR
Fast Healthcare Interoperability Resources (FHIR) has become one of the most common standards for healthcare data exchange. FHIR data typically comes in nested, hierarchical JSON formats, making it challenging to use for data analysis.
Tuva is a standardized, enriched data model that is designed for healthcare research, analysis, and machine learning.
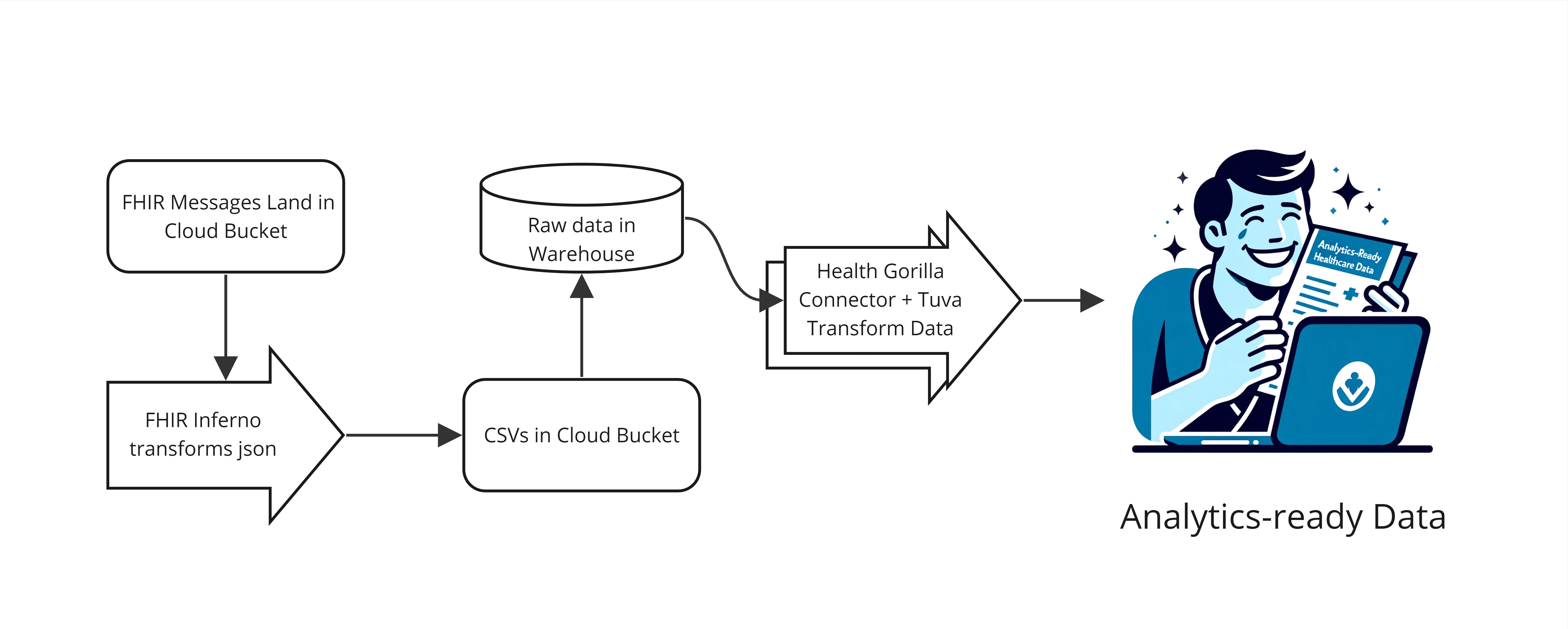
In this guide we describe how to convert FHIR into Tuva, making the data ready for analytics. We call this FHIR Preprocessing. There are 3 tools we use to do this:
- FHIR Inferno: This python utility transforms FHIR in nested JSON into tabular CSVs (i.e. it flattens FHIR).
- Health Gorilla Connector: This dbt project converts flattened FHIR data into the Tuva Input Layer format, preparing it to be transformed by the Tuva repo.
- Tuva: This dbt package transforms data from the Tuva Input Layer format into the Tuva Core Data Model and Data Marts.
We use FHIR data from Health Gorilla in the example that follows. However the tools used can be adapted to work with any sort of FHIR data.
There are a few pre-requisites you'll need in order to follow this workflow:
-
Python 3.x
- External libraries:
- csv output mode: None
- parquet output mode: pandas, pyarrow
- return mode: pandas
- External libraries:
-
dbt 1.3.x or higher
-
A data warehouse supported by dbt
FHIR Inferno
The first step is using FHIR Inferno to flatten nested JSON into tabular CSVs. FHIR Inferno is a Python-based utility designed for this specific purpose. In this section of the guide, we'll walk through the basic setup, configuration, and implementation of the tool.
The core function of FHIR Inferno, the parse function, focuses on flattening FHIR data and creating structured tables. The function applies a config file (an ini formatted configuration detailing the desired table output) to a FHIR resource, transforms it to a tabular format, and either writes the file to disk or returns the object to the caller. The function can also optionally keep track of any file paths that were present in the FHIR resource that weren't known when the config was created, and write those paths to a separate file so the user can adjust configs and reprocess files if needed.
Basic syntax to import and call the function like this:
import parseFhir
parseFhir.parse(r'config/config_Patient.ini', inputPath='FHIR_Input/Patient_0001.json',outputPath='FHIR_output/Patient_0001.csv', missingPath='missing_paths/Patient_0001.csv',outputFormat='csv')
A number of parameters can be passed to the function that affect its behavior:
- configpath: the path to the config file that defines the table structure
- inputPath: the path to the input FHIR resourced
- outputPath: the output path (required for csv or parquet output format)
- missingPath (optional): if present, function will compare paths present in the FHIR resource to those listed in the conifg, and write any that were missing to the file, so they can be reviewed and possibly added to the config
- outputFormat: writes a
csvorparquetto write a file to outputPath, orreturnto return a dataframe - inputFormat:
jsonorndjsondepending on the input file format
The utility comes with out-of-the-box configurations for some common FHIR formats. It also comes with helper scripts that can assist in creating new config files from scratch based on the structure of a set of FHIR resources.
Setup and Configuration
We'll start by downloading the existing configurations for Health Gorilla's flavor of FHIR.
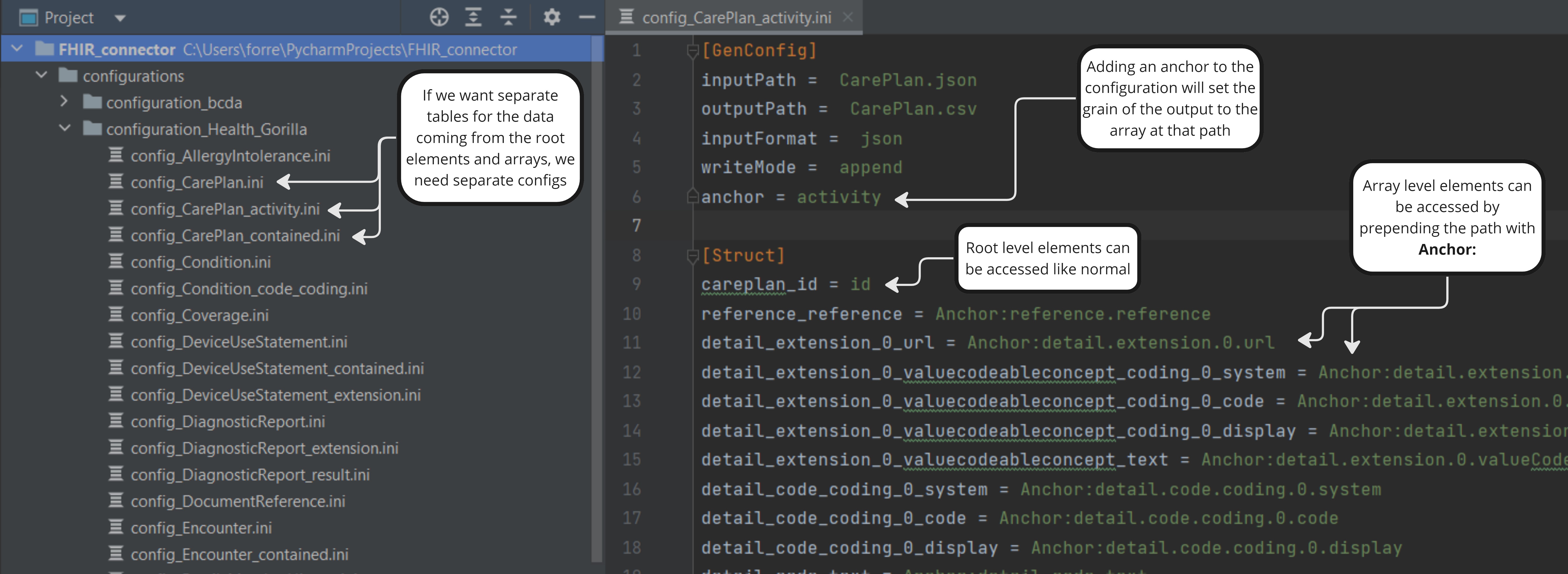
These config files will tell FHIR Inferno what one table's output structure should look like, and where in the FHIR resources to find the data for each column. The config files have a GenConfig section that defines some top level configurations about the transformation, and a Struct section that defines the columns. Note that all of the configurations that can be passed to the function can also be set in the GenConfig section.
The Struct section defines the format of the output table we will be building. Each key in Struct will be a new column in the output, and the value will be a dot-notated JSON path to location in the FHIR resources where the data should be pulled from.
Sometimes we want to build a table at the grain of an array inside a FHIR resource, rather than at the grain of the resource itself. We can provide an anchor key in the GenConfig section that points to the array, and the output will be at that grain. anchor must be set in the config file and can't be passed as a parameter. Once an anchor is set, you can access the anchor elements in by prepending your paths in the Struct section with Anchor:. Root level elements can still be accessed like normal.
If we want multiple tables built off of one FHIR resource, we need multiple config files for that resource, one for each output table. For example, the CarePlan resource can have an "activity" and a "contained" element that are both arrays, and if we want a table for each built, we'll need separate config files for each one.
FHIR Inferno also has some additional basic functionality to do things like apply logic to the JSON paths, perform basic string functions, or write the input filename or date time processed to the output tables. FHIR Inferno can also compare the elements that are present in a FHIR resource to elements that were known when the config file was created and write any missing paths to a CSV, so that a user can be aware of
any new paths or structure changes and take any appropriate actions necessary. For this functionality, it uses additional root_paths or anchor_paths, and an optional ignore_paths section in the config file to identify the know columns. See the
readme for a more detailed breakdown on each section as well as the functions available.
In addition to the core function, FHIR Inferno contains various helper scripts to help with the setup, configuration, and various processing of FHIR resources. It contains scripts to analyze a batch of resources and create configuration files, scripts to build configuration files, and scripts on examples on how to fully implement the solution.
Implementation
Once we have the transformation configs built, it's time to put FHIR Inferno into action. In this example we want the tabular data FHIR Inferno create to be delivered to an AWS S3 bucket, so we'll build a lambda function that will process the files. Our function will primarily work by processing all resources for a patient for a particular file type, running once daily and procesing all of the patients who were added in the last day, but it will also be built to be able to process one file at a time when triggered by an S3 event, if we decide to stream the messages in the future.
It's going to use the return outputMode, so we can aggregate the bulk processed files and write the data as parquet to a separate S3 bucket. It will then write the output as parquet to an S3 bucket, and manage the sqs queue if necessary. We will then set up a snowpipe from our output S3 bucket, so files can flow into our Snowflake environment in real time.
We will handle any updates to files by adding a staging model in our dbt project that takes the most recent version of each record based on the filename and processed date.
Lambda Function
import boto3
import os
import parseFhir
import json
import shutil
import logging
import time
import fnmatch
import pandas as pd
import pyarrow as pa
import pyarrow.parquet as pq
from botocore.exceptions import ClientError
logger = logging.getLogger()
logger.setLevel(logging.INFO)
## cleares the temp directory. With multiple lambda invocations, this can contain resources from previous runs
def clear_tmp_directory():
for filename in os.listdir('/tmp/'):
file_path = os.path.join('/tmp/', filename)
try:
if os.path.isfile(file_path) or os.path.islink(file_path):
os.unlink(file_path)
elif os.path.isdir(file_path):
shutil.rmtree(file_path)
except Exception as e:
logger.exception(f'Failed to delete {file_path}. Reason: {e}', exc_info=True)
# function to call the fhir parse
def execute_parse(resourceType,filepath,outfiledir,anchors,par_dir,filename,agg):
# for aggregating files
if agg:
out_path_group = os.path.join(outfiledir, 'parquet_groups', resourceType, par_dir + '.parquet')
os.makedirs(os.path.dirname(out_path_group), exist_ok=True)
# outMissing = os.path.join(outfiledir,'missing_paths',par_dir, filename + '.csv')
outMissing = os.path.join(outfiledir,'missing_paths', resourceType, par_dir + '.csv')
os.makedirs(os.path.dirname(outMissing), exist_ok=True)
dfs = []
for dirpath, dirnames, filenames in os.walk('/tmp/input/'):
for filename in filenames:
logger.debug(f"file oslistdir inside exec: {dirpath} {filename}")
# outMissing = os.path.join(outfiledir, 'missing_paths', par_dir, filename + '.csv')
# os.makedirs(os.path.dirname(outMissing), exist_ok=True)
df = parseFhir.parse(r'config/config_'+ resourceType +'.ini', inputPath=os.path.join(dirpath,filename), missingPath=outMissing,outputFormat='return')
logger.debug(f"DataFrame summary:\n{df.describe()}")
dfs.append(df)
aggregated_df = pd.concat(dfs, ignore_index=True)
# Write aggregated data to Parquet
table = pa.Table.from_pandas(aggregated_df)
pq.write_table(table, out_path_group)
for anchor in anchors:
dfs = []
out_path_group = os.path.join(outfiledir, 'parquet_groups', resourceType + '_' + anchor, par_dir + '.parquet')
for dirpath, dirnames, filenames in os.walk('/tmp/input/'):
for filename in filenames:
outMissing = os.path.join(outfiledir, 'missing_paths', par_dir, filename + '.csv')
os.makedirs(os.path.dirname(outMissing), exist_ok=True)
df = parseFhir.parse(r'config/config_' + resourceType + '_' + anchor + '.ini', inputPath=os.path.join(dirpath,filename),
missingPath=outMissing,outputFormat='return')
dfs.append(df)
aggregated_df = pd.concat(dfs, ignore_index=True)
# Write aggregated data to Parquet
table = pa.Table.from_pandas(aggregated_df)
os.makedirs(os.path.dirname(out_path_group), exist_ok=True)
pq.write_table(table, out_path_group)
# for processing streamed files
else:
outPath = os.path.join(outfiledir,'parquet_files',resourceType, par_dir, filename + '.parquet')
outMissing = os.path.join(outfiledir,'missing_paths',par_dir, filename + '.csv')
os.makedirs(os.path.dirname(outPath), exist_ok=True)
os.makedirs(os.path.dirname(outMissing), exist_ok=True)
parseFhir.parse(r'config/config_'+ resourceType +'.ini', inputPath=filepath, outputPath=outPath, missingPath=outMissing,outputFormat='parquet')
for anchor in anchors:
outPath = os.path.join(outfiledir,'parquet_files',resourceType + '_' + anchor, par_dir, filename + '_' + anchor + '.parquet')
outMissing = os.path.join(outfiledir,'missing_paths',par_dir, filename + '_' + anchor + '.csv')
os.makedirs(os.path.dirname(outPath), exist_ok=True)
os.makedirs(os.path.dirname(outMissing), exist_ok=True)
parseFhir.parse(r'config/config_' + resourceType + '_' + anchor + '.ini', inputPath=filepath, outputPath=outPath,missingPath=outMissing,outputFormat='parquet')
def choose_config(filepath,outfiledir,agg=False):
path_parts = filepath.split(os.sep)
if len(path_parts) >= 2:
# Join the last two parts of the path (the directory and the file name)
par_dir = path_parts[-2]
else:
# Use only the last part of the path (the file name)
par_dir = ''
filename = os.path.basename(filepath)
outMissing = os.path.join(outfiledir,'missing_paths',par_dir, filename + '.parquet')
os.makedirs(os.path.dirname(outMissing), exist_ok=True)
resourceType = os.path.basename(filepath).split('_')[0]
logger.debug(f"\n--Choosing Congid\nFilepath:{filename}\nResource Type:{resourceType}")
if resourceType == 'AllergyIntolerance':
execute_parse(resourceType,filepath,outfiledir,[],par_dir,filename,agg)
elif resourceType == 'CarePlan':
execute_parse(resourceType,filepath,outfiledir,['activity','contained'],par_dir,filename,agg)
elif resourceType == 'Condition':
execute_parse(resourceType,filepath,outfiledir,['code_coding'],par_dir,filename,agg)
elif resourceType == 'Coverage':
execute_parse(resourceType,filepath,outfiledir,[],par_dir,filename,agg)
elif resourceType == 'DeviceUseStatement':
execute_parse(resourceType,filepath,outfiledir,['extension','contained'],par_dir,filename,agg)
elif resourceType == 'DiagnosticReport':
execute_parse(resourceType,filepath,outfiledir,['result','extension'],par_dir,filename,agg)
elif resourceType == 'DocumentReference':
execute_parse(resourceType,filepath,outfiledir,[],par_dir,filename,agg)
elif resourceType == 'Encounter':
execute_parse(resourceType,filepath,outfiledir,['contained'],par_dir,filename,agg)
elif resourceType == 'FamilyMemberHistory':
execute_parse(resourceType,filepath,outfiledir,['condition'],par_dir,filename,agg)
elif resourceType == 'Immunization':
execute_parse(resourceType,filepath,outfiledir,['contained'],par_dir,filename,agg)
elif resourceType == 'MedicationStatement':
execute_parse(resourceType,filepath,outfiledir,['MedicationCodeableConcept_coding','contained'],par_dir,filename,agg)
elif resourceType == 'Observation':
execute_parse(resourceType,filepath,outfiledir,['hasMember','extension','contained'],par_dir,filename,agg)
elif resourceType == 'Organization':
execute_parse(resourceType,filepath,outfiledir,[],par_dir,filename,agg)
elif resourceType == 'Patient':
execute_parse(resourceType,filepath,outfiledir,['address'],par_dir,filename,agg)
elif resourceType == 'Procedure':
execute_parse(resourceType,filepath,outfiledir,['contained','reasonCode'],par_dir,filename,agg)
else:
logging.error('Missed a resource type: ' + resourceType, exc_info=True)
raise
def lambda_handler(event, context):
logger.debug(f"Starting: {json.dumps(event)}")
try:
clear_tmp_directory()
s3_client = boto3.client('s3')
local_input_path = '/tmp/input/'
local_output_path = '/tmp/output/'
os.makedirs(local_input_path, exist_ok=True)
os.makedirs(local_output_path, exist_ok=True)
# S3 event information
input_type = None
if event.get('Records') and event['Records'][0].get('eventSource') == 'aws:sqs':
input_type = 'sqs'
try:
body = event['Records'][0]['body']
receipt_handle = event['Records'][0]['receiptHandle']
event_data = json.loads(body)
except json.JSONDecodeError as e:
logger.error("fError parsing SQS message body: {e}")
raise e # or handle the error as you see fit
else:
input_type = 'trigger'
event_data = event
s3_event = event_data['Records'][0]['s3']
bucket_name = s3_event['bucket']['name']
agg = s3_event.get('agg', False)
prefix = s3_event.get('prefix', 'zzzzz')
pattern = s3_event.get('pattern', '*')
skip_count = s3_event.get('skip', 0)
recursion_depth = s3_event.get('recursion_depth', 0)
file_key = s3_event.get('object', {}).get('key')
logger.info(f"\n--Processing\nBucket:{bucket_name}\nprefix:{prefix}\npattern{pattern}\nskip_count:{skip_count}\nrecursion_depth:{recursion_depth}\nagg:{agg}\nfile_key:{file_key}")
if agg:
# Check the recursion depth
if recursion_depth > 25:
logger.error("Maximum recursion depth reached.")
return {
'statusCode': 400,
'body': json.dumps('Maximum recursion depth reached')
}
# List and process files
s3_resource = boto3.resource('s3')
bucket = s3_resource.Bucket(bucket_name)
processed_files_count = 0 # Counter for files processed after skipping
total_files_count = 0 # Total files examined
local_input_file = None # Initialize the variable with a default value
for obj in bucket.objects.filter(Prefix=prefix):
if fnmatch.fnmatch(obj.key, pattern):
local_input_file = os.path.join(local_input_path, obj.key)
if obj.key.endswith('/'): # Skip 'folders'
logger.debug(f"Skipping 'folder' key: {obj.key}")
continue
if total_files_count < skip_count:
total_files_count += 1
logger.debug(f"\n--Skipping\nObject key: {obj.key}, Local input file path: {local_input_file}")
continue # Skip this file
logger.debug(f"\n--Downloading\nObject key: {obj.key}, Local input file path: {local_input_file}")
try:
os.makedirs(os.path.dirname(local_input_file), exist_ok=True)
s3_client.download_file(bucket_name, obj.key, local_input_file)
except Exception as e:
logger.exception(f"Error occurred while processing {obj.key}: {e}", exc_info=True)
continue
processed_files_count += 1
total_files_count += 1
# Check if limit is reached
if processed_files_count >= 1000:
# Trigger next Lambda function
sqs = boto3.client('sqs')
new_event = {
"Records": [
{
"s3": {
"bucket": {
"name": bucket_name
},
"object": {
"key": obj.key
},
"prefix": prefix,
"pattern": pattern,
"skip": skip_count + 1000,
"recursion_depth": recursion_depth + 1,
"agg": True
}
}
]
}
message_body = json.dumps(new_event)
logging.info(f'\n--Invoking sqs\nPrefix: {prefix}\nPattern: {pattern}\nRecursion Depth: {recursion_depth}')
sqs.send_message(
QueueUrl="https://sqs.us-east-1.amazonaws.com/123456789012/health_gorilla_queue",
MessageBody=message_body
)
break
if local_input_file is None:
logger.warning("No valid input file found.")
return {
'statusCode': 204,
'body': json.dumps(f'\nNo valid input file found.\nBucket:{bucket_name}\nprefix:{prefix}\npattern{pattern}\nskip_count:{skip_count}\nrecursion_depth:{recursion_depth}\nagg:{agg}\nfile_key:{file_key}')
}
choose_config(local_input_file, local_output_path, True)
## single file processing
else:
local_input_file = '/tmp/input/' + file_key
try:
os.makedirs(os.path.dirname(local_input_file), exist_ok=True)
s3_client.download_file(bucket_name, file_key, local_input_file)
choose_config(local_input_file, local_output_path, False)
processed_files_count = 1
total_files_count = 1
except Exception as e:
logger.error(f"Error occurred while processing {file_key}: {e}")
# Upload processed files to S3
upload_processed_files(local_output_path, s3_client,recursion_depth)
if input_type == 'sqs':
try:
sqs = boto3.client('sqs')
sqs.delete_message(
QueueUrl="https://sqs.us-east-1.amazonaws.com/123456789012/health_gorilla_queue",
ReceiptHandle=receipt_handle
)
except Exception as e:
logger.error(f"Failed to remove message from queue: {e}")
logger.info(f"\nSuccessfully processed {processed_files_count} files. Total files examined: {total_files_count}\nBucket:{bucket_name}\nprefix:{prefix}\npattern{pattern}\nskip_count:{skip_count}\nrecursion_depth:{recursion_depth}\nagg:{agg}\nfile_key:{file_key}")
return {
'statusCode': 200,
'body': json.dumps(f'Processed {processed_files_count} files. Total processed: {total_files_count}')
}
except Exception as e:
logger.error("An error occurred", exc_info=True)
raise
def upload_processed_files(local_output_path, s3_client,recursion_depth):
output_bucket_name = 'output-bucket'
for dirpath, dirnames, filenames in os.walk(local_output_path):
for filename in filenames:
file_path = os.path.join(dirpath, filename)
if os.path.isfile(file_path):
relative_path = os.path.relpath(file_path, start=local_output_path)
if recursion_depth > 0:
base, ext = os.path.splitext(relative_path)
modified_relative_path = f"{base}_{recursion_depth}{ext}"
output_file_key = os.path.join('FHIR_Output', modified_relative_path)
else:
output_file_key = os.path.join('FHIR_Output', relative_path)
output_file_key = output_file_key.replace(os.path.sep, '/')
s3_client.upload_file(file_path, output_bucket_name, output_file_key)
Health Gorilla Connector
Now that our data is transformed to a tabular format and loaded to our data warehouse, we can use the Health Gorilla Connector to convert the data into the Tuva Input Layer.
We'll start by cloning the Health Gorilla Connector. Our goal is to transform our data into the Tuva Input Layer, so that we can run Tuva on the data.
Health Gorilla Configs configures the data into the format expected by the Health Gorilla Connector. If our data is in another format, we'll have to build some custom models to get our data into the Tuva Input Layer format.
Once we have our own version of the connector cloned locally, all of the configurations we need to make will be in the dbt_project.yml file:
- if our profile name is anything other than
default, we need to change theprofile:configuration to match what we set in our profiles.yml - we need to set the
input_databasevar to the name of the database where our raw Health Gorilla data is - we need to set the
input_schemavar to the name of the schema where our raw Health Gorilla data is
We can also use this opportunity to set any normal dbt configurations we want, such as the output database or schema and any custom documentation pages, etc.
Tuva
Once configured, we can pull in the latest versions of the Tuva package with a dbt deps, and then run all of our models with dbt build. This will transform the data into the Tuva Input Layer and then the Tuva Core Data Model, Data Marts, as well as load the Reference Data, Terminology, and Clinical Concept Library.